ABOUT
We use molecular dynamics (MD) simulations and related theoretical and computational methods to investigate the self-assembly, phase behavior and interaction networks of a range of soft-matter biological systems.
In MD simulations, a complex molecular assembly is modeled by a set of interacting particles, whose evolution in time and space is calculated by numerical integration of Newton’s second law. The method is rigorously based on the laws of statistical mechanics, and allows calculation of bulk thermodynamic properties while simultaneously providing atomic-scale resolution of molecular behavior that cannot directly be obtained from analytical measurements alone.
News
Popular Media Coverage
- On magic mushrooms: https://sciencenews.dk/en/psychedelic-effects-of-psilocin-at-the-molecular-level
- More on magic mushrooms: https://www.sdu.dk/en/om_sdu/fakulteterne/naturvidenskab/nyheder-2023/psilocin-research
- Videnskab.dk: Mechanism of the Umami Taste Sensation
- Videnskab.dk: An antipyshotic repurposed as an anticancer drug
- CSCS: The stoichiometry of the gastric H, K, ATPase
Research Areas
- Annexin-mediated Membrane Remodeling
- Ion Pumps
- Accelerating Simulations
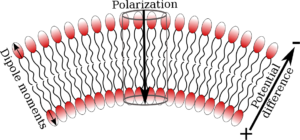
- Flexo-Electricity in Membranes
- Drug-Membrane Interactions
- Oxidized Phospholipids
- Magic Mushrooms and Psychedelics
project Examples
Ion Pumps
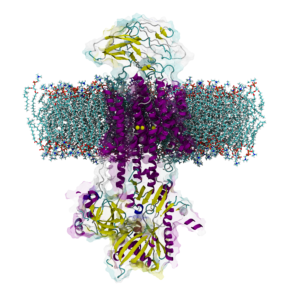 We investigate the molecular basis of regulation, transport and neurological diseases in Ion Pumps. We work with the Na, K ATPase, the gastric H, K ATPase, and the novel and the unique KdpFABC channel-pump bacterial transport complex. Our simulations are in collaboration with leading experimental groups in Japan, USA, Australia and Europe.
We investigate the molecular basis of regulation, transport and neurological diseases in Ion Pumps. We work with the Na, K ATPase, the gastric H, K ATPase, and the novel and the unique KdpFABC channel-pump bacterial transport complex. Our simulations are in collaboration with leading experimental groups in Japan, USA, Australia and Europe.
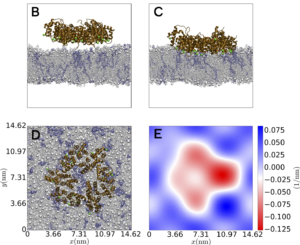
Membrane Repair Mechanisms in Cancer Cells
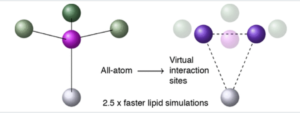
Method Development
Electromechanical Coupling in Lipid Membranes

